4.9 PROJECT MENU
- 4.9.1 about the project menu (general comments)
- 4.9.2 edit project composition
- 4.9.3 merge project sequences (and close)
- 4.9.4 project file lists
- 4.9.5 project statistics
- 4.9.6 find identical sequences in project
- 4.9.7 sequence list form
- 4.9.7.1 file menu (list form)
- 4.9.7.2 save tab-delimited list
- 4.9.7.3 export to excel spreadsheet
- 4.9.7.4 view menu (list form)
- 4.9.7.5 format menu (list form)
- 4.9.7.6 options menu (list form)
- 4.9.7.7 align menu (list form)
- 4.9.7.8 entrez menu (list form)
- 4.9.7.9 find menu (list form)
- 4.9.7.10 refresh menu (list form)
- 4.9.7.11 user comments (edited description lines)
 4.9.1 about the project menu
4.9.1 about the project menu
Under this menu are collected functions/facilities designed to edit project composition and to display a list os sequences included in the project. The latter form provide extensive options for customising the way project sequences are listed. In addition to a simple list containing "raw" sequence data (length, orientation, checksum etc.) it is possible to use information contained in the sequence header to list the project sequences. The sequence list form ![]() is explained in detail below.
is explained in detail below.
 4.9.2 edit project composition
4.9.2 edit project composition
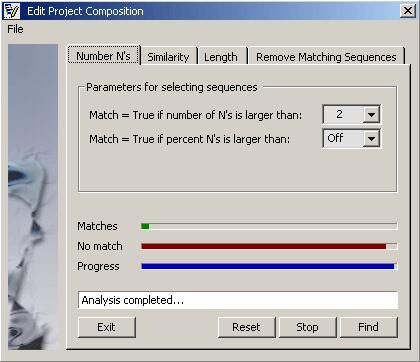
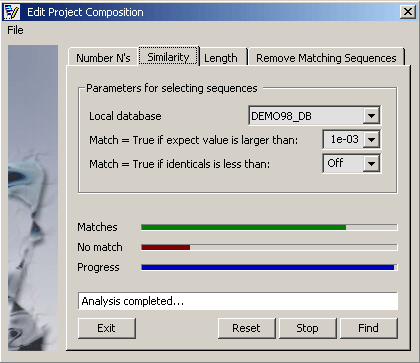
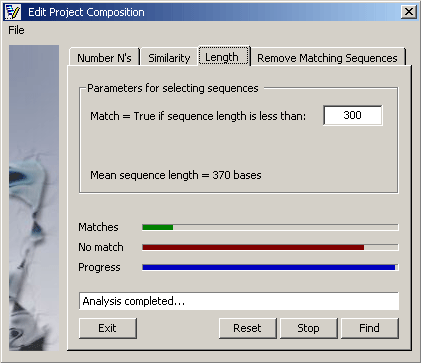
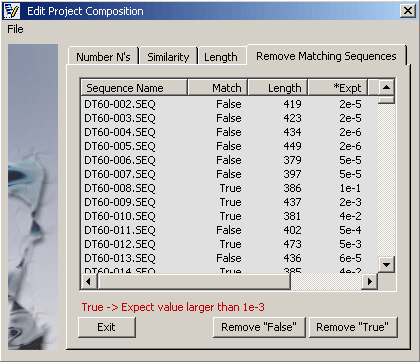
This form contains three functions for finding and removing sequences from the project: Bad quality sequences
with many ambigous positions (N's), sequences similar to those contained in a specified local database (for example
containing a selections of vector sequences) and sequences shorter than a specified minimum length.
Parameters for finding low quality sequences.
Parameters for finding similar sequences.
Parameters for finding short sequences.
Result list. The Match column indicates whether or not the specified criteria were met for all
project sequences. Click Remove "False" to close sequences not meeting the criteria
or Remove "True" to close sequences meeting the criteria.
 4.9.3 merge project sequences
4.9.3 merge project sequences
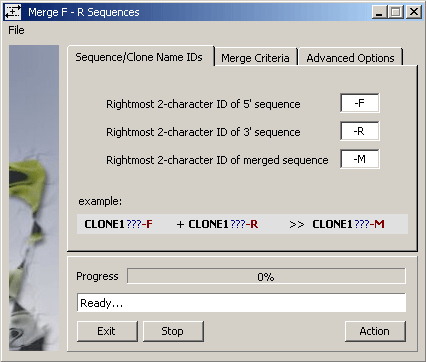
This function also appears under the Compose menu. It is included here because it allows
you to replace mergeable forward and reverse sequence pairs originating from the same plasmid / insert by
their merged sequence - closing the forward and reverse sequences after successfully creating the merged sequence.
Consult the Compose menu for more details.
 4.9.4 project file lists
4.9.4 project file lists
This menu includes various options for printing and saving project sequence file lists. As described
in detail under the File menu the plp and psp
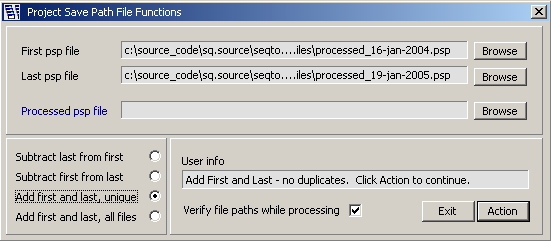
files can be used to open the project from a list of full file paths. The psp file editor allows you to combine
two psp files in various ways.
The psp file editor and its options.
 4.9.5 project statistics
4.9.5 project statistics
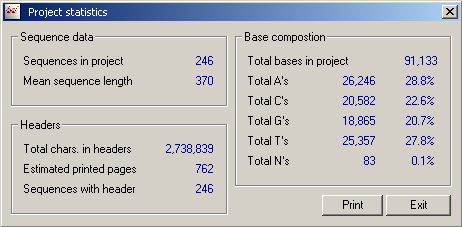
This function calculates and lists various quantitative information for the project sequences.
 4.9.6 find identical sequences in project
4.9.6 find identical sequences in project
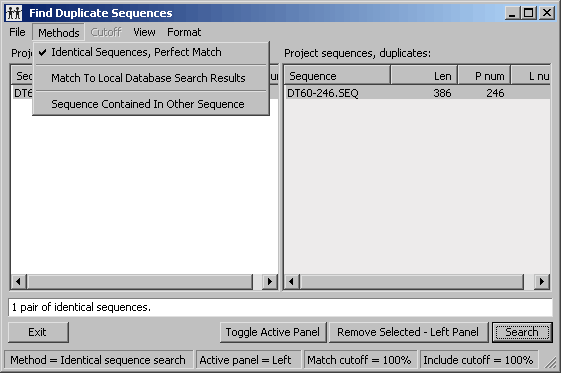
A quick way to find and close duplicate (identical or similar) sequences in the project.
 4.9.7 sequence list
4.9.7 sequence list
The sequence list form ![]() is
rather complex with a very large number of options for displaying the project sequences based on available information
in sequence headers. Many of the options require that you have performed one or more blast search and retrieved
relevant supplementary information from Entrez (e.g. full genbank records for the best matches of blast searches).
The list options (other than listing plain sequence data) also require that you include the information you wish to
use in the sequence list in the virtual header
is
rather complex with a very large number of options for displaying the project sequences based on available information
in sequence headers. Many of the options require that you have performed one or more blast search and retrieved
relevant supplementary information from Entrez (e.g. full genbank records for the best matches of blast searches).
The list options (other than listing plain sequence data) also require that you include the information you wish to
use in the sequence list in the virtual header ![]() .
.
The facilities are explained in some detail below to present the possibilities for extracting and displaying information
stored in sequence headers. The row of icons on the right hand panel of the form provides direct access to most of the
functions and options also included in the menus.
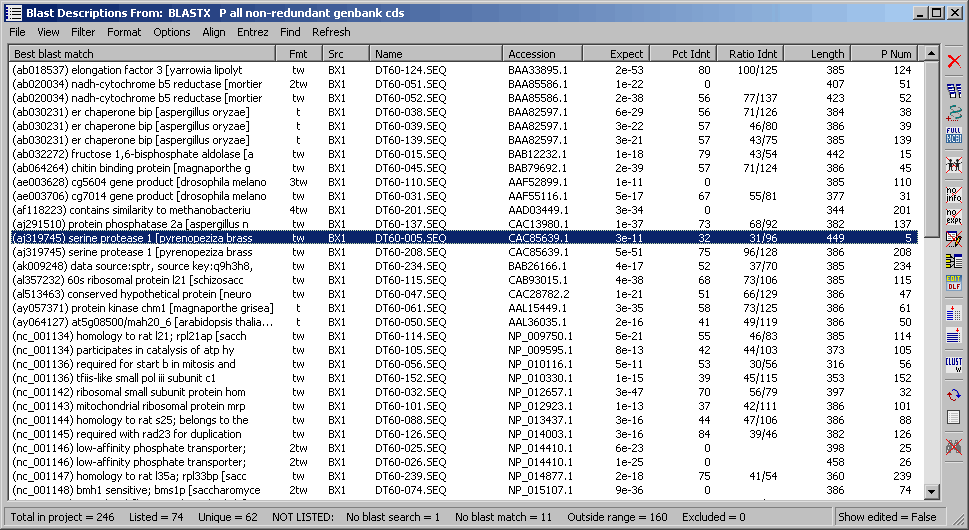
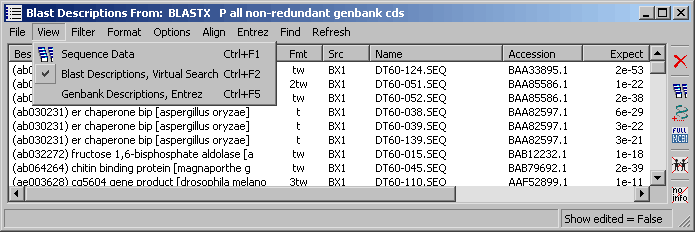
This screenshot below shows the project sequences listed according to the search result of a blastx search
on genbank section: all non-redundant genbank cdc selected as the virtual blast search in
the Header Compose form. All information in the table is
derived from the blast description and alignment sections of the sequence header. Other options for data display are
included under the View menu the available options reflecting information included in the virtual blast search.
Example: If you include Genbank records, best match in the virtual blast search an option to use this
information for displaying project sequences in iscluded under the View menu, whereas this option is not
available if Genbank records are omitted from the virtual blast search.
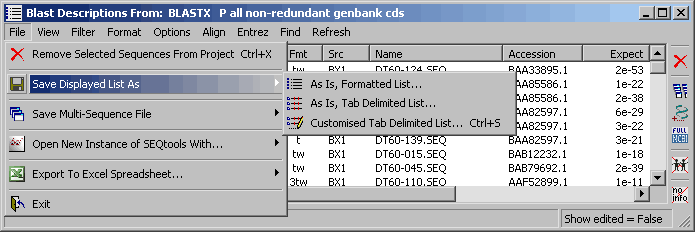
4.9.7.1 File menu - The File menu includes various options for exporting the project sequence list. For all
functions the exported list is the currently displayed project sequence list. The choises for each export option
varies, but are self explanatory. Briefly you can export highlighted lines or lines not highlighted, include information
in individual columns, etc.
4.8.7.2 Tab-delimited sequence list for saving the project sequence list. - This facility allows you to compose and save a tab-delimited project sequence list including selected sequences from the project sequence list.
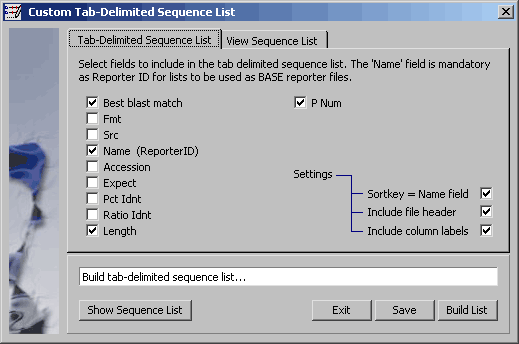
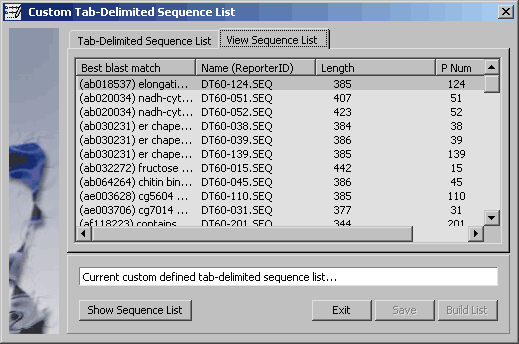
Project sequence list consisting of the checked items.
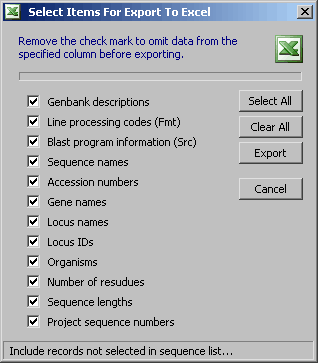
4.9.7.3 Export to Excel spreadsheet - Options form for exporting the project sequence list to an Excel
spreadsheet.
4.9.7.4 View menu - The content of the View menu relflects the header information available for the project
sequences. Enabled menu items causes an update of the project sequence list reflecting the source of information.
4.9.7.5 Format menu - The Format menu items provide access to several functions/settings affecting the
line format:
- Build Virtual Blast search enables you to change the content of the virtual blast search.
- Edit FastA Definition Lines is a editor for composing/editing the content of definition lines in fasta formatted
sequences.
- Format Description Lines, described in detail under the Preferences menu, is an advanced filter for
customising / filtering description lines from blast search results.
- Line Formatting Off turns off line filtering.
- Edit Description Lines opens the User Comments form to enable you to add verious comments and to create your own Genbank descriptions (only enabled when the project sequence list displays Genbank records). See below for details.
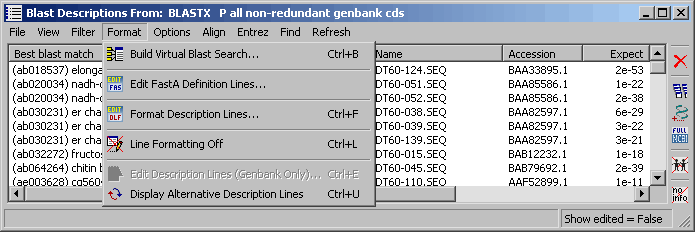
4.9.7.6 Options menu - A quick way to alter the width of the description column of the list.
4.9.7.7 Align menu - The project sequence list provides direct access to alignment by Clustal. Simply select
the sequences you want to align and click ClustalW (SEQtools user interface) or ClustalX (with its own unser interface and
many more options):
4.9.7.8 Entrez menu - When the Launch Entrez With Accession Number On Shift-Click is enabled you can
highlight a line in the project sequence list containing an accession number (i.e., list display by blast
search results or by Genbank records) and shift-click to launch retrieval of the highlighted sequence in a
web browser from Entrez. webbrowser with the se:
4.9.7.9 Find menu - The search option allows you to search either description lines
only or all other
fields (and not description lines). Hits are highlighted in the
project sequence list.
4.9.7.10 Refresh menu - These options are used to rebuild/update the project sequence list. In most cases the
list is updated automatically but some changes may require manual action. It is also possible to invert selections
and to remove highlights (not the sequences).
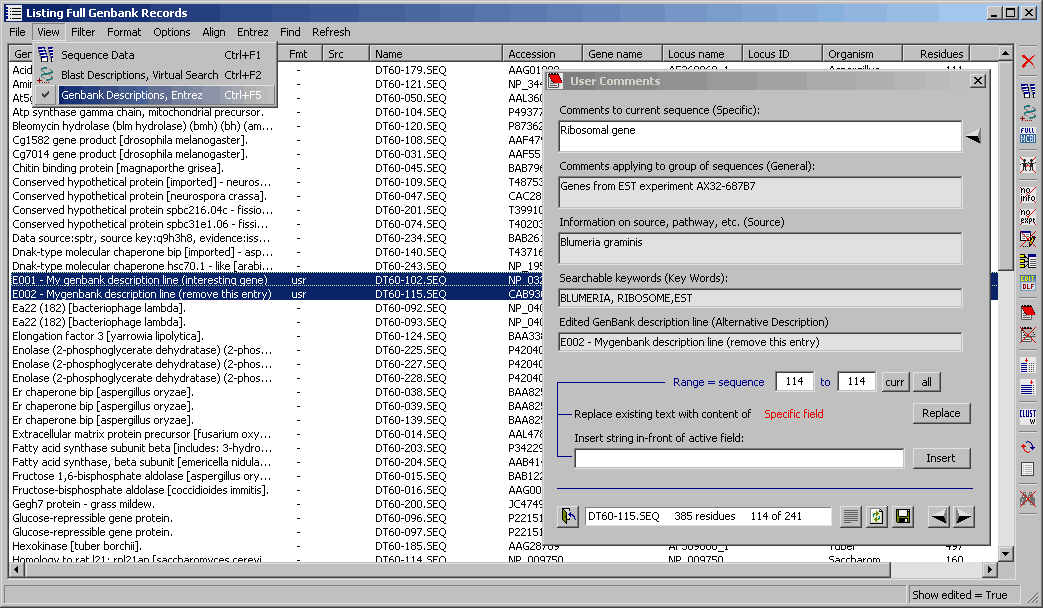
4.9.7.11 User edited Genbank information - Sometimes you may want to create an alternative description
line to a given gene, add a comment to the gene (or to all sequences in the project), to add key words to
facilitate searching headers or grouping sequences or to add your initials (for example) in front of the descriptions.
The User Comments form ![]() is intended
for these purposes.
is intended
for these purposes.
The highlighted menu item in the form below allows you to use the original Genbank description lines or those
created by yourself. The highlighted lines in the sequence list have been changed to illustrate this facility.
Note that this option is only available for Genbank descriptions.
© 2002-2010S.W. Rasmussen (revised: )